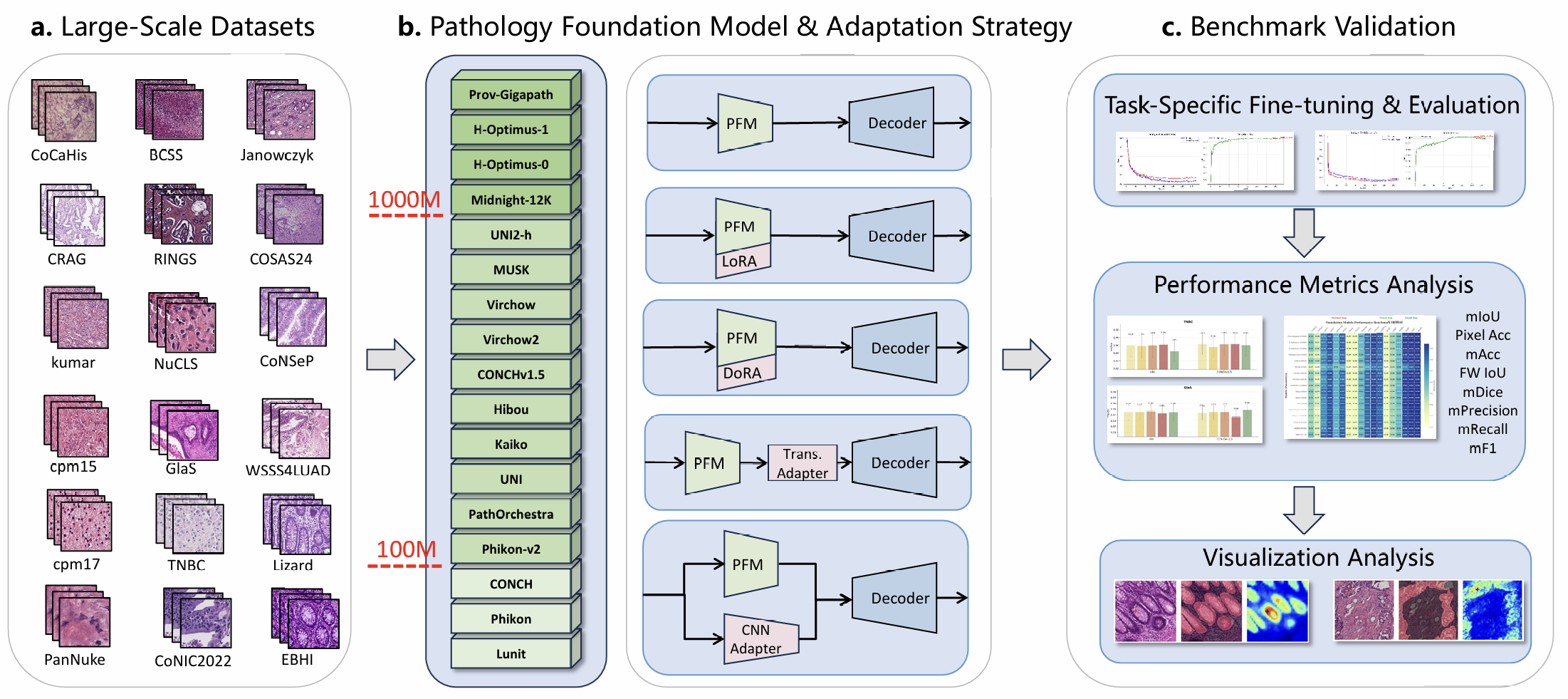
Figure 1. Overview of PFM-DenseBench: A unified benchmark for evaluating Pathology Foundation Models on dense prediction. The framework comprises dataset curation, model and strategy evaluation, and benchmark validation.
To What Extent Do Token-Level Representations from Pathology Foundation Models Improve Dense Prediction?
Bridging the gap between foundation model evaluation and clinical dense prediction needs
While most PFM benchmarks focus on image-level classification, clinical diagnosis relies on precise pixel-level segmentation. We evaluate PFMs where it matters most.
Unified protocols across 18 datasets enable fair comparisons. No more scattered benchmarks with incompatible setups and metrics.
Discover which fine-tuning strategy works best for your task. From frozen encoders to CNN adapters, we reveal what actually drives performance.
From nuclei to glands to tissue regions—our benchmark spans the full spectrum of biological scales relevant to computational pathology.
Does bigger always mean better? Our findings challenge conventional wisdom and reveal the true drivers of dense prediction performance.
All code, configs, and containers are publicly available. Bootstrap confidence intervals ensure statistically rigorous comparisons.
Comprehensive evaluation framework covering diverse datasets, models, and adaptation strategies

Figure 1. Overview of PFM-DenseBench: A unified benchmark for evaluating Pathology Foundation Models on dense prediction. The framework comprises dataset curation, model and strategy evaluation, and benchmark validation.
CoNIC2022, PanNuke, CPM15, CPM17, CoNSeP, Kumar, NuCLS, Lizard, TNBC
GlaS, CRAG, RINGS
BCSS, CoCaHis, COSAS24, EBHI, WSSS4LUAD, Janowczyk
UNI, UNI2-h, Virchow, Virchow2, Phikon, Phikon-v2, H-Optimus-0/1, Prov-GigaPath, Hibou-L, Kaiko-L, Lunit, PathOrchestra, Midnight-12k
CONCH, CONCHv1.5, MUSK
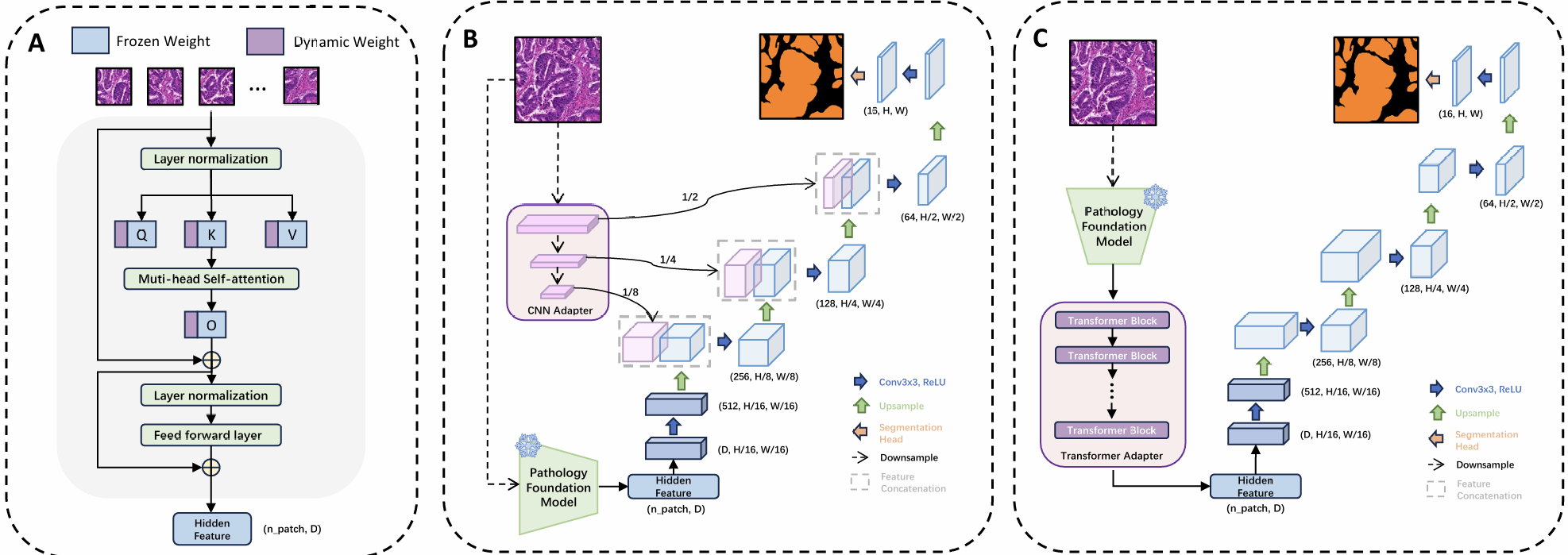
Figure 2. Architecture of adaptation strategies. (A) Low-Rank Adaptation (LoRA/DoRA). (B) CNN Adapter with multi-scale convolutional branches. (C) Transformer Adapter with additional transformer blocks.
Comprehensive results across all datasets and models (mDice metric)
Best mDice score achieved on each dataset with the corresponding model and adaptation method
| Dataset | mDice (SOTA) | Best Model | Method |
|---|---|---|---|
| Loading data... | |||
Average rank across all 18 datasets and 5 adaptation methods (lower is better)
| Rank | Model | Avg. Rank (lower is better) |
|---|---|---|
| Loading data... | ||
If you find PFM-DenseBench useful in your research, please cite our paper
@misc{chen2026extenttokenlevelrepresentationspathology,
title={To What Extent Do Token-Level Representations from Pathology Foundation Models Improve Dense Prediction?},
author={Weiming Chen and Xitong Ling and Xidong Wang and Zhenyang Cai and Yijia Guo and Mingxi Fu and Ziyi Zeng and Minxi Ouyang and Jiawen Li and Yizhi Wang and Tian Guan and Benyou Wang and Yonghong He},
year={2026},
eprint={2602.03887},
archivePrefix={arXiv},
primaryClass={eess.IV},
url={https://arxiv.org/abs/2602.03887},
}